Where on the Phylogenetic Tree Should the Origin of the Supercell Appear?
- Original Knowledge domain Clause
- Unenclosed Entree
- Published:
How to Read a Biological process Sir Herbert Beerbohm Tree
Evolution: Education and Outreach volume 3,pages 506–519 (2010)Advert this article
Abstract
It has been ended 50 days since Willi Hennig proposed a new method for determining kindred relationships among species, which he called phylogenetic systematics. Many people, however, still approach the method warily, upset that they will have to grapple with an overwhelming number of new terms and concepts. In fact, reading material and understanding phylogenetic trees is really not difficult at all. You only need to hear three spick-and-span language, autapomorphy, synapomorphy, and plesiomorphy. All of the different concepts (e.g., ancestors, monophyletic groups, paraphyletic groups) are familiar ones that were already part of Darwinian evolution before Hennig arrived on the fit.
Dan Brooks and I Blackbeard a biodiversity course (EEB 265) to second year students at the University of Toronto. The entire course is structured around a phylogenetic framework. We commence with the big, albeit simplified, tree of the Metazoa, past crop our way from sponges to snakes, focusing on the characters that bind groups together and the characters that make for each one group unique. If we are doing our job correctly, our students should equal able to answer the favourable questions—what is this animal (how set you know)? What does information technology do? What makes it particular? What aspects of its biology make it vulnerable to anthropogenic intercession? Since all of the students had already taken a lab in first year biology covering the basics of phylogenetics, we assumed that we wouldn't need to review phylogenetic methodology in our biodiversity run. It didn't take long for us to realize that our Assumption was naïve; by the clock time many of the students had arrived in EEB 265, they had already hit the delete clit next to "phylogenetics" in their brain. It is always humbling to (ray)discover that not everyone shares your views about the things in life that are interesting and consequential!
Back to the drawing board. Unmatchable of the major problems with teaching a course about metazoan diversity is that you simply don't give enough time to cover all of the groups. The high thing we wanted to do was to ritual killing biology-based lectures for a discussion about theory. So, the dispute was simple: design a lecture that would, in 50 minutes, teach students how to understand what a phylogenetic tree was telling them. Information technology wasn't our intention to teach students how to make trees, just how to read them. This paper is supported that take to task.
The word "phylogeny" is a combination of two Greek words, phyle (tribe—in particular, the largest political subdivision in the ancient Athenian state of matter [www.yourdictionary.com; World Wide Web.etymonline.com]: another word we get from this is "phylum") and geneia (origin [www.etymonline.com]: some other word we get from this is "gene"). IT was coined by the developmental life scientist Ernst Ernst Heinrich Haeckel in 1866 and then championed by Darwin in his famous put to work, On the Origin of Species (beginning with the 5th edition in 1869). Some biologists tied the theme of "phylogeny"—the origin of groups—to evolution. Biological process trees are thus simply diagrams that render the origin and evolution of groups of organisms.
Although you might not know it, we are all familiar with the idea of phylogenetic trees. People have been making such trees for decades, substituting the word, "family" for "phylogenetic" (Fig. 1). Just as someone the great unwashe in a family over generations are connected by bonds of "blood" (the process of replica that produces offspring), individual species are connected by biological process ties (biological processes like natural selection and geologic processes such as continental swan or a river changing course that make species). In this signified, speciation (the output of new species) = reproduction (the production of new individuals). In separate run-in, we are all, from members of the same syndicate to members of the same species, connected by genes.
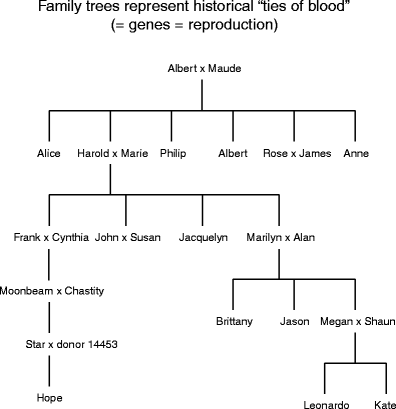
Family Sir Herbert Beerbohm Tree for an interesting group of citizenry. In phylogenetic terms, family trees (genealogies of people) = phylogenetic trees (genealogies of species)
Category trees tend to glucinium haggard as if they were hanging upside down, like a flock of grapes. Phylogenetic trees are depicted within reason other than. Imagine that you are belongings the genealogy for the big cats shown in Fig. 2a. Forthwith, flip information technology sideways (rotate 90° levorotatory) and you have the image shown in 2b. Rotate this image yet some other 90° counterclockwise, smooth it out, and you have the image shown in Fig. 2c (this tree shape was the one used aside Darwin in Happening the Origin of Species). The important thing to remember is that all three depictions are expression exactly the same thing about the relationships among species of big cats. How you prefer to draw your phylogenetic trees depends, in split, on personal orientation—some people find it easier to read 2b, others prefer 2c.
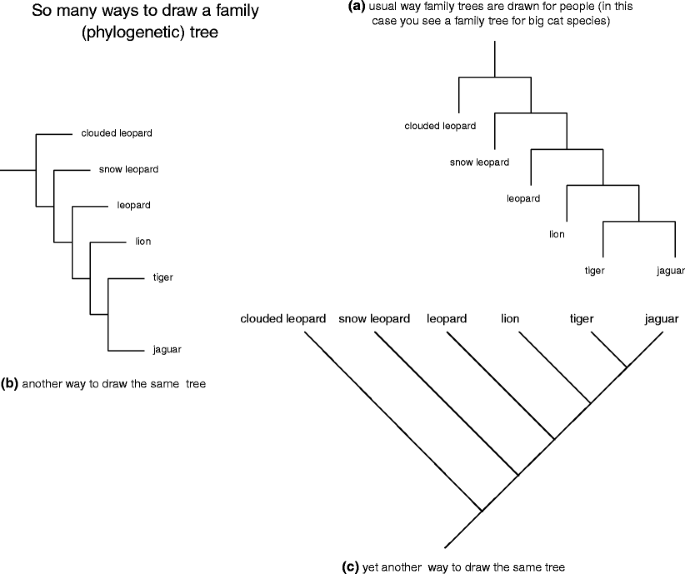
a–c So many ways to thread a family/phylogenetic tree for the genus Panthera
Phylogenetic trees are reconstructed away a method named "phylogenetic systematics" (Fig. 3). This method acting clusters groups of organisms together based upon shared, unique characters called synapomorphies. For example, you share the mien of a moxie with cats, but not with butterflies. The presence of a backbone thus allows us to hypothesize that anthropomorphous beings are more closely related to cats than they are to butterflies (Fig. 4a); cats and people both have a backbone, butterflies are spineless Footnote 1. Not entirely characters are synapomorphies. Some traits, called plesiomorphies, are distributed by all the members of a group. Returning to our tree, we see that cats, people, and butterflies all have DNA (Fig. 4b). The presence of DNA allows us to hypothesize that these trio species are all part of the homophonic group, but information technology does not tell U.S.A anything about how those species are age-related to one another. Think of it this way: my cognomen tells me that I am part of the McLennan clan. If I foregather someone known as Jessie McLennan, I lie with we are related for some reason, but I haven't any idea whether she is a long lost full cousin or someone from a many distant limb of the family tree. The final term you need to bang is autapomorphy—traits that are only plant in one extremity of the group. For example, butterflies hind end be important from cats and people because they have an exoskeleton made out of chitin (a punk, waterproof first derivative of glucose). Autapomorphies help US identify a particular species in a group but, like plesiomorphies, they state us nothing or so relationships inside the chemical group. Overall these three types of characters can constitute likened to the story of Goldilocks: plesiomorphies are too hot (too widespread), autapomorphies are overly low temperature (too restricted), and synapomorphies are to perfection (for deciding biological process relationships).
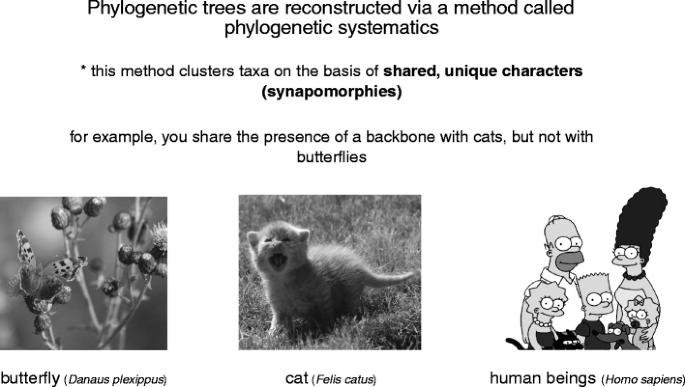
The basis of organic process systematics
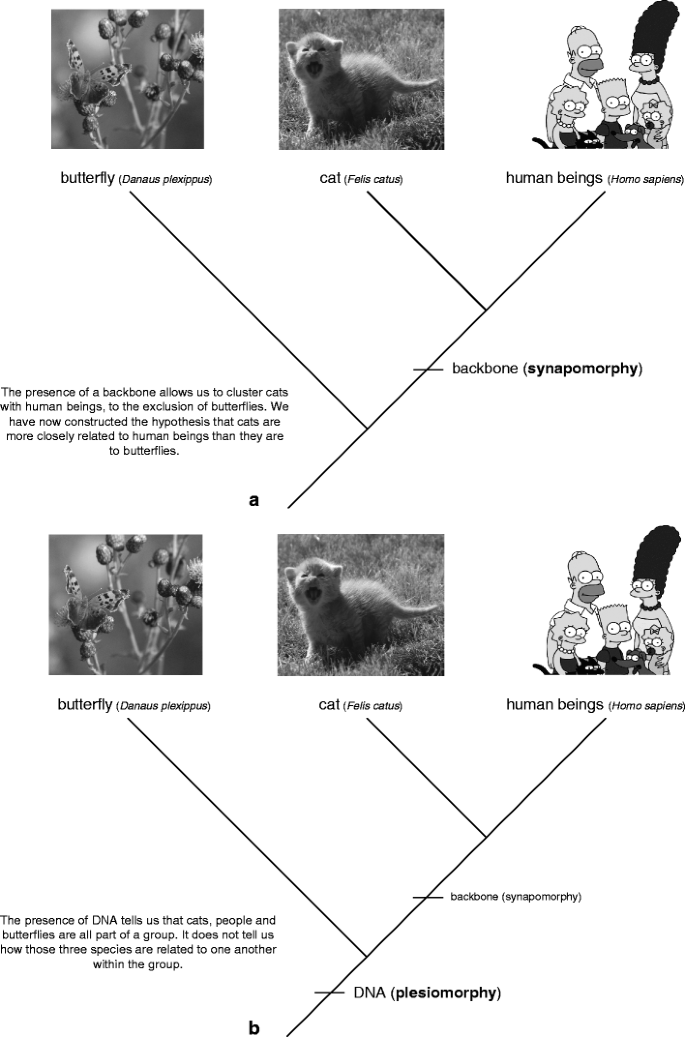
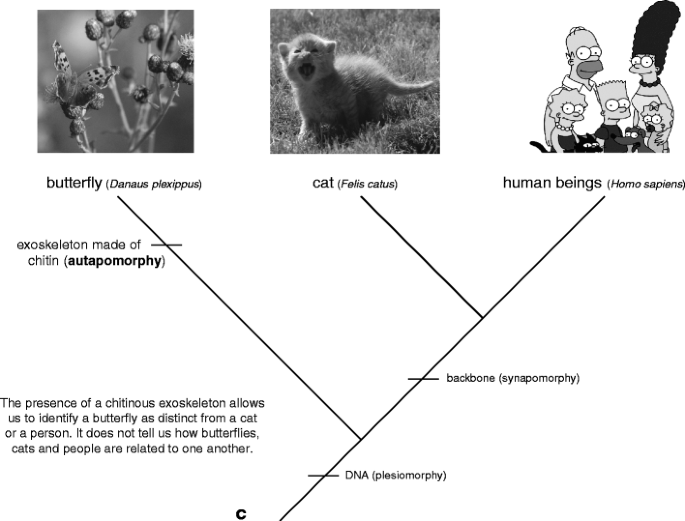
Characteristic types of characters on a phyletic tree. a a synapomorphy; b a plesiomorphy; c an autapomorphy
Enough of characters for the moment; back to the trees themselves. Why behave the branches connected a tree have names (e.g., Leo, tiger, etc.), while the lines connexion distinct branches together do not (Libyan Islamic Fighting Group. 5)? This is because these lines symbolise ancestors. An ancestor is a species that has undergone a speciation issue to produce descendant species. The ancestor usually "disappears" in the process of speciation. Does this awful that the root goes extinct?
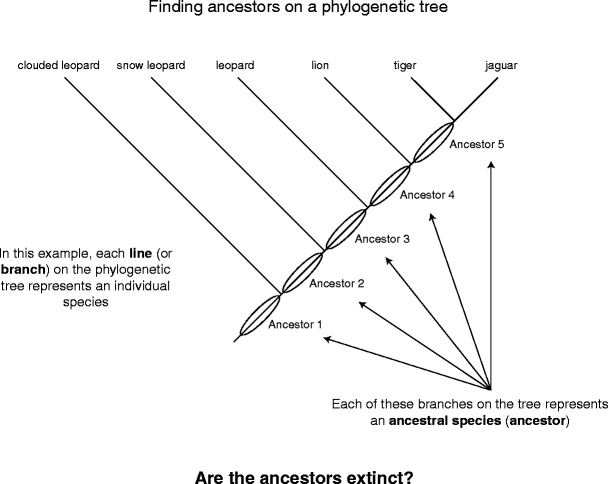
Determination ancestors on a phylogenetic tree
In order to answer this, we must do roughly time traveling carrying a integer device that records everything we see (Fig. 6). Imagine you go under back 10,000,000 years, past stop, intrigued by an interesting species of lizard with red muscae volitantes all over its back (species A). After a spell, you decide to move forward in time five million years some then catch again. You search around and discover deuce radical lizard species, combined with blue spots on its back (species B), and the other with red grade insignia (species C), but species A is nowhere to equal seen. Did information technology survive extinct? You looking back over your digital recording of those fin million years and discover that species A split into two groups, which became different in some ways from cardinal another through time. In evolutionary terms, species A is an ascendant (ancestor 1) and species B and C are its descendants. Fast forward to today (with more digital physical to watch) and you find trey species of lizard: your old friend the blue spotted lizard (species B) and two new lizards (descendants of species C, the reddened stripy lounge lizard), one with blue stripes (species D) and the other with a solid black hindermost (species E). Today, so, thither are only three species of lizard alive. You no more see either of the ancestors (the ruby-red sullied and red striped lizards), but we static prove them on the phyletic tree.

Traveling backward in time to expose ancestors
The answer to our original question "did the ancestor conk out extinct?" is thence No! In many cases, the ancestor is subdivided and the begotten (genetic) information encompassed inside the ancestor is passed on to the descendent species. Over time, the descendants change and become different in whatever ways from each otherwise and from the ancestor, patc retaining some things in common (for example, all of our lounge lizard species have a back). This is evolution.
So what really counts as extinguishing? Extinction is the loss of natural selective information—the physical red ink of a species. For example, consider a easy phylogenetic tree of the dinosaurs (Fig. 7). All of the groups on dotted branches are extinct—none of the species in those groups exist on this planet any longer (Jurassic Common notwithstanding), which substance that entirely of the information that was incomparable to each of those groups has been lost. The only grouping that managed to avoid extinction was Aves (or birds)—avian species are the conclusion remaining dinosaurs.
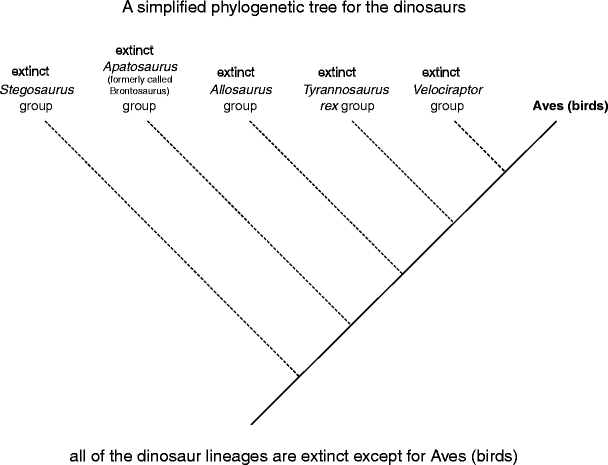
Factual extinctions. Groups depicted with dotted lines are extinct so totally of the genetic, morphological, physiological, ecological, and behavioral traits that are unique to from each one grouping have been lost to the biosphere
Fine, let's take what we take up learned almost ancestors and clustering groups supported shared, unique characters (synapomorphies) and use that to trace the selective information restrained within a phylogenetic tree. Here is a tree depicting the relationships among living members of the Amniota, a large group of vertebrates that includes to the highest degree of the animals with which you are familiar (Libyan Islamic Grou. 8). You already know that the names of species, operating theatre groups of species, are written across the tips of the branches on the shoetree. The close thing you need to know is that characters are depicted at their point of origin on a phylogenetic corner. So, on this tree you can see that (1) the amnionic egg originated in ancestor 1 and was passed on to altogether of its posterity (mammals, ancestor 2, turtles, ancestor 3, ancestor 4, crocodiles, birds, ancestor 5, tuataras, and lizards positive snakes). In evolutionary terms, the sac ball is a incomparable trait that is distributed lone by ancestor 1 and all of its descendants; (2) a special character of scrape protein (β keratin) originated in ancestor 2 and was passed on to altogether of its posterity (turtles, ancestor 3, ancestor 4, crocodiles, birds, ancestor 5, tuataras and lizards plus snakes). β ceratin is a unique trait shared by the group called "Reptilia"; and (3) a frail tail originated in ancestor 5 and was passed on to all of its descendants (tuataras, lizards asset snakes). A breakable tail is a unique trait shared aside members of the group tuataras + lizards + snakes.
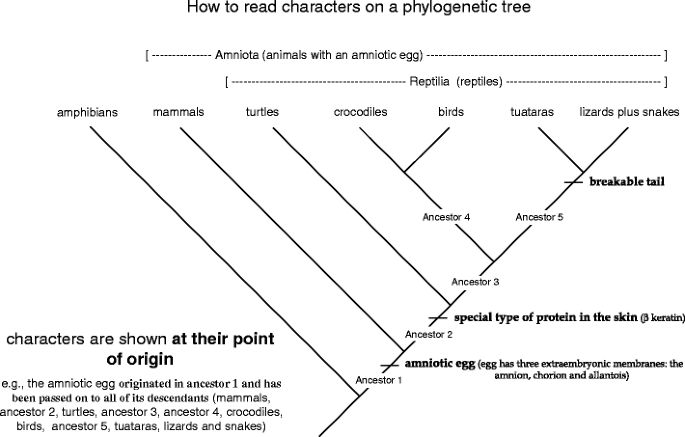
How to read characters on a phylogenetic tree
In point of fact, every being is a complex mosaic of thousands of traits. If you don't believe this, sit down and list totally of the traits that make you, you. In addition to the obvious things like eye color and hair color, don't forget the fact that you deliver RNA, DNA, individual cells, an anterior and posterior remainder, a skull, jaws, bone, arms and legs, come from an amniotic nut, stimulate three maraca in your labyrinth, were suckled on milk produced in mammary glands, suffer an opposable thumb, and no tail. In otherwise words, when you face at a biological process tree, you volition see that all of the branches take up at least unmatched, and more likely some, characters on them (the slash marks on Libyan Fighting Group. 9a). Because of this, it is oftentimes difficult to really label all of the traits connected a tree because information technology's visually distracting. A shorthand method acting has been developed to deal with this job: draw the tree showing the relationships among the groups (Fig. 9b) and list the synapomorphies for each branch elsewhere in a table. Then again, if you are interested in one or many particular traits, you can foreground them along the phyletic tree without viewing complete the other characters. For instance, if you wanted to discuss the evolution of mammals, you could show off the amniote tree and highlight just the synapomorphies for the mammals (e.g., three tympanic cavity bones: Common fig tree. 9c). Remember, this is just shorthand!
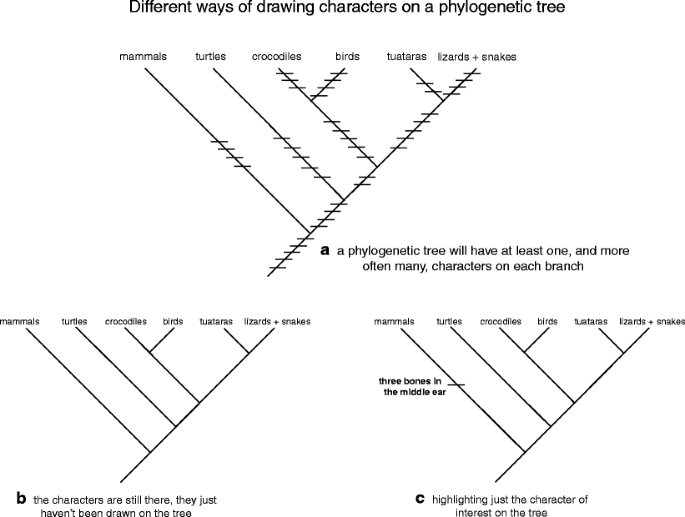
a–c Representing characters on a phylogenetic tree
There is one last thing about characters that is important to realise: characters are not static things. They evolve through time. In other words, a "synapomorphy" may non "look the same" altogether species that have information technology. So, for object lesson, consider the stapes, one of the three finger cymbals in your middle capitulum that are answerable for transferring sound waves from the eardrum to the membrane of the internal ear. This soft bone has a long, complicated, and fascinating organic process history. To understand that history, we must journey rachis many of hundreds of millions of years to the line of descent of the Deuterostomes, a large group that includes the Echinodermata (sea star and their relatives), Hemichordata (twist-like, marine creatures), and Chordata (lancelet + tunicates + Craniata [organisms with skulls]). The ancestor of this large group had many slits in its pharynx (named splanchnic arches) that were involved with filter feeding. Time passed and cartilaginous rods providing support for the arches appeared, were divided and varied. The upper section of the secondly viscus curve rod cell is the focus of our tale (FIG. 10). As we move forward still further in time, this character undergoes various geophysics and point modifications; fundamentally, it becomes larger, more robust, and involved in bearing the jaws (at which point IT is named the hyomandibula), changes from gristle to bone, then begins a step-by-step reduction in size, disengages from the jaw/cheek area, and moves into the tympanum (at which point it is known as the stapes). Gross then, the upper portion of the 2nd internal organ arch—hyomandibula—stirrup is the same social organization that has had both its shape and routine modified over hundreds of millions of years. So although the bearing of a "cartilaginous perch in the 2nd visceral arch found in the throat part" may be a synapomorphy for the Craniata, you won't find that exact structure in whatever quadrupedal animals. Rather, what you will incu is the modification of that gristly perch, the stapes. The continued evolution of a peculiar case past its point of stemma is called an organic process transformation series.
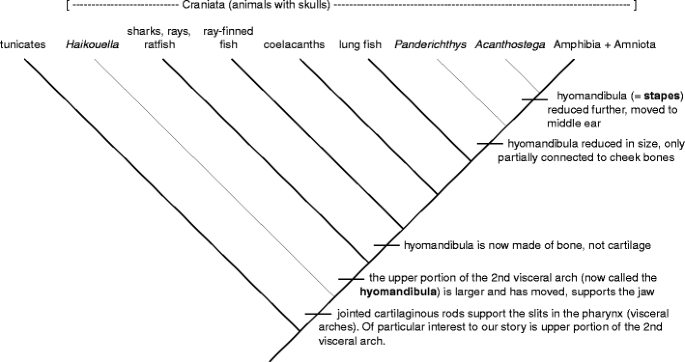
Synapomorphies are non static; they may continue to acquire. Changes in the character "upper portion of the second illogical patronizing" [hyomandibula, stapes] are traced on the phylogenetic corner for the Chordata (animals with notochords). Both the story and the phylogenetic corner have been substantially simplified to emphasize the idea of character stemma and modification kind of than the finer details of character evolution. Names in italics refer to extinct species known from fossils. Line drawings and photographs of different structures and species can be found easily on the entanglement
The next affair that students of phylogenetics have to jazz is how to recognize different kinds of groups of organisms. There are two general types of groups, one "well behaved" and the other "bad".
Let's begin with "the good," a monophyletic mathematical group (Fig. 11). The word "monophyletic" is a combination of two Greek row, monos (unvarying) and phyle (tribe). It was coined by our old acquaintance Ernest Haekel, who, as you call up, too invented the word phylogeny. A monophyletic group includes an ancestor and all of its descendants. It is known by the presence of shared out, specific characters (synapomorphies). Each phylogenetic tree contains as galore monophyletic groups as there are ancestors. For example, looking at the tree diagram in Fig. 11, we can place five monophyletic groups, only two of which are shown on Common fig tree. 12 (I'll allow it adequate to you to discover the other terzetto).
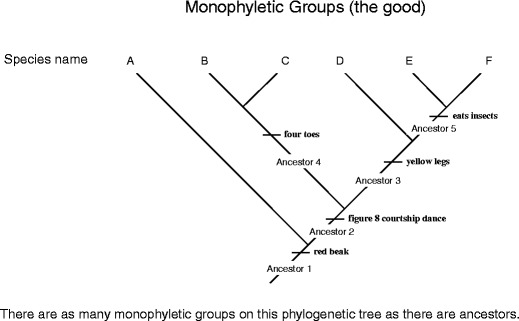
Distinguishing monophyletic groups
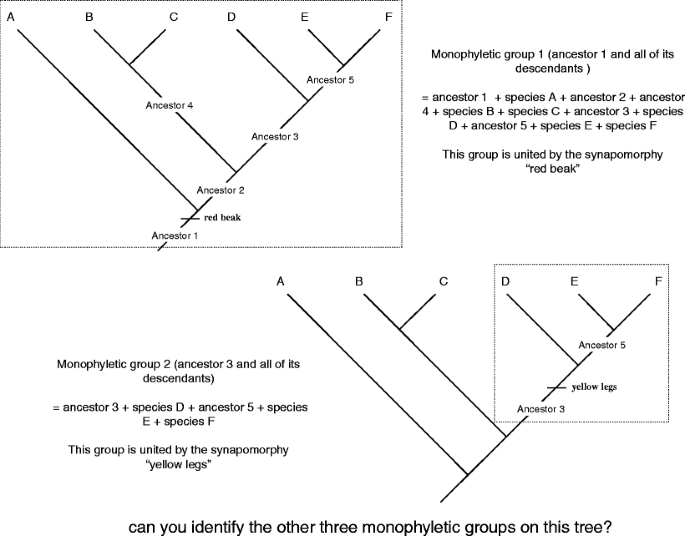
Ii of the five monophyletic groups on the divinatory tree
Straightaway onto "the bad." The word "paraphyletic" is, once again, a combining of two Geek words, para (near) and phyle (kin group), and then the implication is that the whole tribe is not stage (Fig. 13). Paraphyletic groups include an ancestor but not all of its descendants. Happening this hypothetical tree, species C has been eliminated from the group, even though it is a descending of ancestor 1 just like the rest of the species. Paraphyletic groups are tough because they misinform us about how characters germinate and how species are related to one another. E.g., net ball's consider the astronomical tree for the Amniota and highlight the "old" Reptilia, one of the most famous paraphyletic groups (Fig. 14). Even today people calm down speak about three distinct classes, the reptiles, the birds, and the mammals. When you look at this figure, what is untimely about the class Reptilia, the way it is drawn?
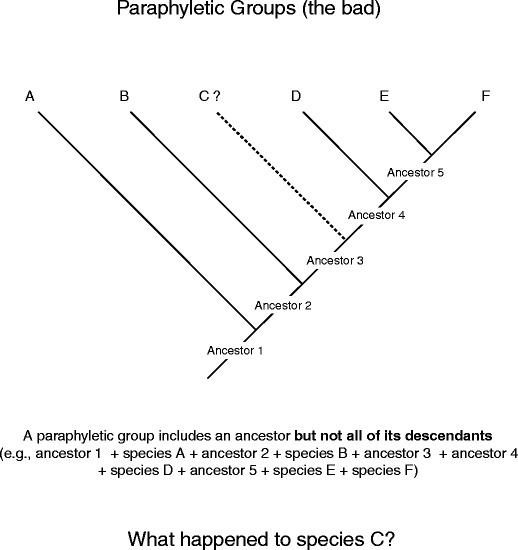
Identifying paraphyletic groups
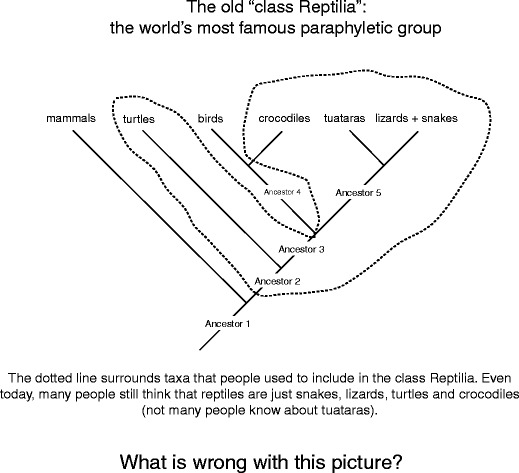
The most famous paraphyletic group, the reptiles
Right! In (Fig. 15) Ancestor 2 is the ancestor of all the reptiles only, American Samoa highlighted on this figure, the Reptilia does not include all of ancestor 2's posterity; ancestor 4 and the birds wealthy person been distant from the group. The exclusive right smart to make the Reptilia a monophyletic group is to redefine the term to include crocodiles, turtles, tuataras, lizards, snakes, and birds. In the preceding, birds were not thoughtful to be reptiles because they are warm-full-blood (in fact, they were often grouped with mammals because of that trait). But phyletic studies have incontestible that birds are indeed reptiles because they share many morphological, behavioral, and unit characters with unusual reptilian species in miscellaneous (synapomorphies originating in ancestor 2; e.g., β keratin), and they share many characters with crocodiles in particular (synapomorphies originating in antecedent 4; e.g., holes in the skull just before of the eyes).
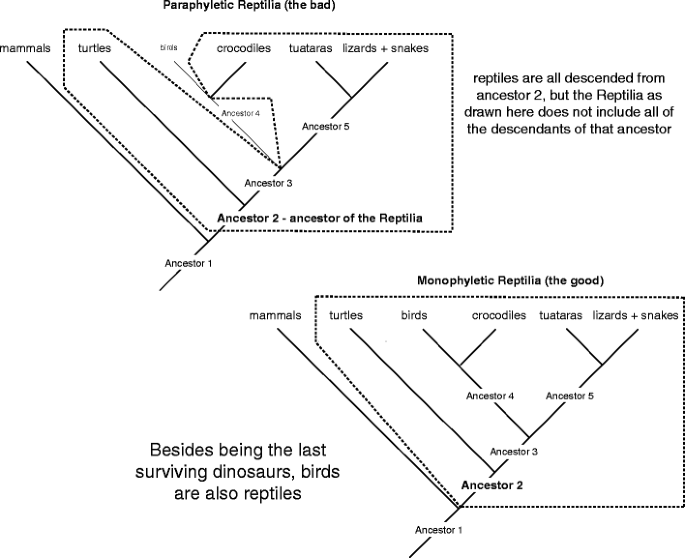
How to make the Reptilia monophyletic
Wherefore is information technology life-or-death to consume monophyletic groups? Say you wanted to work how red fuzz appeared in your family. What would represent your chances of tracking down your original red-comate ancestor if none records were kept about the union between your great-eager-great-great grandfather Sven and his Irish bride Maggie? Missing information creates problems for any research, be it genealogical or organic process, and paraphyletic groups are missing information. In evolutionary terms, monophyletic groups are "real" biological units; that is, they are the product of descent with modification (an ancestor and all of its descendants) and as so much can be used to study the evolutionary processes that produced them. Paraphyletic groups, on the else hand, are the product of "human error" arising from sketchy or flawed selective information (e.g., piteous descriptions of characters). Using such groups to study organic process processes will direct us along misleading and confusing pathways.
Why do we use phylogenetic trees? At that place are umpteen ways to answer this question (and many papers/books written about it), simply the most general answer is that trees summarize valuable information about the evolution of organisms that allows us to empathize them better. For example, here's the kinsfolk shoetree for the Superfamily Hominoidea, the group that includes us and whol of our nighest relatives (Libyan Fighting Group. 16). When you count at the dispersion of characters on this tree you can see that a act of traits we associate only with human beings, such as hunting, infanticide, tool making, self-awareness, and language, originated long before Homo sapiens. Put differently, human beings are not arsenic specific as you might think. If we want to see how and wherefore those traits evolved, we must study their reflexion and occasion in ourselves and in our relatives. So much information from just one organic process tree!
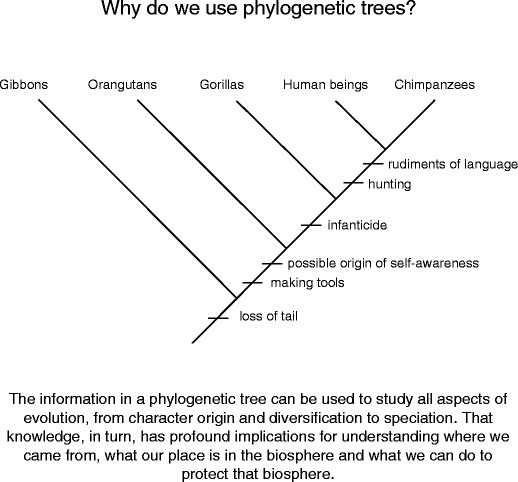
Exploitation a phylogeny to study ourselves
Notes
- 1.
Exposure of crowned head mas courtesy of Daniel Brooks; icon of the Simpsons from Simpsons wallpaper (web.simpsonstrivia.com.ar/simpsons-wallpaper.htm); Kitten is Taala (which means "wind" in the language of the Nuxalk Land people, Bella Coola, BC)
Author information
Affiliations
Corresponding author
Rights and permissions
Open Admittance This is an open access article apportioned under the terms of the Fictive Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, dispersion, and replication in any medium, provided the original author(s) and source are attributable.
Reprints and Permissions
About this article
Where on the Phylogenetic Tree Should the Origin of the Supercell Appear?
Source: https://evolution-outreach.biomedcentral.com/articles/10.1007/s12052-010-0273-6
0 Response to "Where on the Phylogenetic Tree Should the Origin of the Supercell Appear?"
Post a Comment